(2025-Jan-13) Working on the data masking project has been a long journey; writing about it will take some time. I’m fine if you’re not a big fan of long reads and prefer scrolling, skipping, or engaging in shorter, bite-sized learning experiences. I will keep this writing for myself as a chronicle of my journey along a path that may seem overwhelming. However, the experience throughout this journey and the joy of reaching the final destination will be hard to forget.
When I think of data masking, or the term "masking" in particular — the intention to “hide” something from someone comes to mind. I picture a man in a mask, presumably to conceal his identity, or an image blurred beyond recognition. In the world of computer data, masking represents the process of obfuscating, redacting, altering, hiding, encrypting, anonymizing, or substituting sensitive information that requires protection.
One way to protect sensitive information from end users in a database is through dynamic masking. In this process, the actual data is not altered; however, when the data is exposed or queried, the results are returned with modified values, or the actual values are replaced with special characters or notes indicating that the requested data is hidden for protection purposes.
Data Masking Workflow
Ok, let’s put on a developer’s hat. You’ve been assigned to create a solution to mask tables in a relational database. You know which tables are involved, and someone has informed you that not all columns in those tables need to be masked. You’ve also heard about different methods to mask or alter the data.
Your first instinct as a developer might be to run a few UPDATE SQL statements to permanently change and protect the PII data. It all sounds good, but it lacks a controlled masking workflow, and relying on manual efforts as a routine practice is not ideal. This approach can lead to inconsistencies and fails to ensure the reliable protection of sensitive data across all environments.
Not a problem. Let’s move forward with the understanding that we can automate those UPDATE statements and incorporate them into several scripts or a more sophisticated data transformation solution. You’ve been well-trained and can choose the tools that suit you best.
The process remains the same: references to tables, their columns, and different masking methods are embedded in your crafted solution. The number of scripts or data flow pipelines corresponds to the number of tables that need protection. I’m hysterically filled with joy at the sight of explicit table column names hardcoded in my solution.
I’m being sarcastic because I’ve seen this approach many times in other people’s work, and I’m guilty of it myself doing similar things in the early days of my IT career.
Why this approach is bad: any future schema changes will require additional effort to update the data flow pipelines. If I need to replicate the same generic data update mechanism for multiple tables, I’ll have to apply it to every single table. Now imagine being deep into development and discovering a mistake that affects all your database table routines. At that point, your maintenance efforts can quickly spiral into a nightmare, and screaming for help might not offer much relief.
What do we do? We need to build a solution that allows for configuring tables, table columns, masking functions or methods, and assigning those masking functions to specific table columns. This data masking solution should also be scalable to support many tables – whether it's dozens, or even hundreds. Your database and business requirements for data protection and governance will define the limits.
To keep it short – this data masking solution was built using the Azure Databricks platform to mask 290 Oracle database tables, with more than 1,200 specific columns assigned to 10 different masking methods. These methods ranged from simple data substitution and lookup dictionary redactions to regulated randomization.
Why was Databricks chosen? Simple, it’s a scalable cloud data platform that can handle data flow solutions for a single table or scale up to many more as needed. We only pay for the actual workload, with no additional licensing fees for third-party solutions. Plus, Databricks supports multiple programming languages: Python, for example, and when it comes to data masking, Python naturally comes to mind.
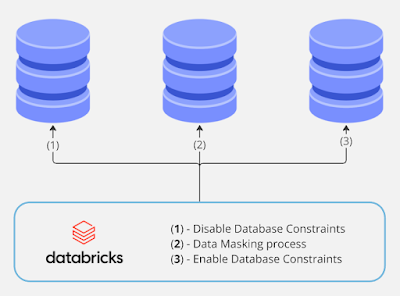
Following this order in the data masking process ensures data integrity and consistency in a relational database. Disabling database constraints (Step 1) prevents errors during masking when constraints like foreign keys or unique indexes might block updates. The masking process (Step 2) then modifies sensitive data as required. Finally, re-enabling constraints (Step 3) restores the database's structural safeguards, ensuring that all data adheres to the original relational rules after masking is complete.
Masking Scope Configuration
Let's have a look at the actual data masking scope configuration.
CREATE TABLE [cfg].[MaskingConfigDatabase] ( [DatabaseID] INT IDENTITY(1,1) NOT NULL PRIMARY KEY CLUSTERED, [WorkspaceName] VARCHAR(50) NULL, [HostName] VARCHAR(50) NULL, [Port] INT NULL, [DatabaseName] VARCHAR(50) NULL ) ON [PRIMARY]; GO
A single entry in the configuration table represents the Oracle database configuration for masking. The table includes critical information such as WorkspaceName, HostName, Port, and DatabaseName.
CREATE TABLE cfg.MaskingConfigTable ( ID INT IDENTITY(1,1) NOT NULL PRIMARY KEY CLUSTERED, DatabaseName VARCHAR(50) NULL, SchemaName VARCHAR(50) NULL, TableName VARCHAR(50) NULL, PrimaryKeyColumn VARCHAR(50) NULL, ColumnName VARCHAR(50) NULL, MaskingRule VARCHAR(100) NULL, MaskingType VARCHAR(50) NULL ); GO
The masking process for a target database is configured through the table [cfg].[MaskingConfigTable], which defines the specific tables and columns targeted for masking. Each entry outlines key attributes such as SchemaName, TableName, and PrimaryKeyColumn, ensuring that data masking operations are precisely scoped at the table and column level. The ColumnName field identifies sensitive data elements that require masking, while the MaskingRule specifies the approach used, such as data_redaction_simple for straightforward text redaction or data_lookup_substitution for city and postal code substitution. The MaskingType further categorizes the nature of masking, including text, email, city, and postal code, aligning with the sensitivity of the data.
Here is the list of currently scoped and custom-developed masking methods using Python notebooks in Databricks. Please note that this list may differ from what is needed for your particular case. You are free to develop your own masking functions as needed.
Cloning Data for Masking
Working with real data involves the feeling, “Oh, what if I make a mistake and do something wrong with the actual data? What should I do to correct it, and how can I prevent it from happening again?”
This natural concern leads us to clone and make copies of the tables required for masking in a separate schema. We don’t clone the entire database, only the objects scoped for masking.
Additionally, to protect against data corruption during schema cloning within the database, a decision was made to create a copy of all the scoped tables in a separate, secure Azure Blob Storage as well.
Python Function for Table Cloning: Initial Stage
def dm_flow_table_backup(database_name, table_name, operations_timestamp, backup_types, df_target_table_data, df_target_table_metadata): try: run_logging_to_blob_history('Info', f'dm_flow_table_backup({database_name})', None, None, None, None, None, 'Started') target_table_name = table_name source_table_name = f"{target_table_name.split('.')[0]}_src.{target_table_name.split('.')[1]}" if 'database' in backup_types: update_masking_table_status(table_name, str(operations_timestamp), "CloneDBStatus", "Started") dm_flow_table_backup_database(target_table_name, source_table_name) update_masking_table_status(table_name, str(operations_timestamp), "CloneDBStatus", "Completed") if 'file_storage' in backup_types: update_masking_table_status(table_name, str(operations_timestamp), "CloneStorageStatus", "Started") dm_flow_table_backup_blob(database_name, target_table_name, operations_timestamp, df_target_table_data) update_masking_table_status(table_name, str(operations_timestamp), "CloneStorageStatus", "Completed") run_logging_to_blob_history('Info', f'dm_flow_table_backup({database_name}, {backup_types})', None, None, None, None, None, 'Completed') except Exception as e: error_message = str(e) run_logging_to_blob_history('Error', f"Error occurred while cloning to either Azure Blob or Database: {error_message}", 'Dataset Cloning', None, None, None, None, 'Failed') run_logging_to_sql_history() raise
Python Function for Table Cloning: Database
def dm_flow_table_backup_database(table_name_to_clone, table_name_to_create): try: # Log the start of the operation run_logging_to_blob_history('Info', f'dm_flow_table_backup_database({database_name}:{table_name_to_clone}): Database schema creation', 'Dataset Cloning', database_name, None, None, None, 'Started') # Split schema and table name schema_name, table_name = table_name_to_create.split('.') # Check if the table already exists table_exists_query = f""" SELECT COUNT(*) AS TABLE_COUNT FROM all_tables WHERE owner = '{schema_name.upper()}' AND table_name = '{table_name.upper()}' """ mss_user, mss_password, jdbcUrl, jdbc_driver_class = dm_oracle_connection() df = spark.read \ .format("jdbc") \ .option("url", jdbcUrl) \ .option("dbtable", f"({table_exists_query})") \ .option("user", mss_user) \ .option("password", mss_password) \ .option("driver", jdbc_driver_class) \ .load() df_count = int(df.collect()[0]['TABLE_COUNT']) if df_count > 0: # Table exists run_logging_to_blob_history('Info', f"dm_flow_table_backup_database({database_name}:{table_name_to_clone}): Table {table_name_to_create} already exists, skipping creation", 'Dataset Cloning', database_name, None, None, None, 'Skipped') print(f"{datetime.now()} Table {table_name_to_create} already exists, skipping creation.") return # Exit the function # Table does not exist; proceed with creation dm_execute_oracle_command(f"CREATE TABLE {table_name_to_create} AS SELECT * FROM {table_name_to_clone}") # Log successful completion run_logging_to_blob_history('Info', f'dm_flow_table_backup_database({database_name}:{table_name_to_clone}): Database table creation', 'Dataset Cloning', database_name, None, None, None, 'Completed') print(f"{datetime.now()} Table '{table_name_to_create}' has been cloned in the database successfully.") except Exception as e: # Handle exceptions and log the error error_message = str(e) run_logging_to_blob_history('Error', f"dm_flow_table_backup_database({database_name}:{table_name_to_clone}): Database table creation: {error_message}", 'Dataset Cloning', database_name, schema_name, None, None, 'Failed') run_logging_to_sql_history() raise
Python Function for Table Cloning: Azure Blob Storage
def dm_flow_table_backup_blob(database_name, table_name, operations_timestamp, df_target_table_data): try: # Constants SMALL_DATA_THRESHOLD = 1000 LARGE_DATA_THRESHOLD = 10000 records_per_partition = 1000000 df = df_target_table_data output_blob_path = MOUNTPOINT + "/mss/backup/" + operations_timestamp + "/" + table_name # Count the number of records in the DataFrame record_count = df.count() print(f"{datetime.now()} Number of records in the DataFrame: {record_count}") # Define the number of records per partition # Calculate the number of partitions, ensuring at least one partition num_partitions = max(math.ceil(record_count / records_per_partition), 1) print(f"{datetime.now()} Number of partitions in the DataFrame: {num_partitions}") # Adjust partitions if record_count < SMALL_DATA_THRESHOLD: df = df.coalesce(1) # Small DataFrame else: df = df.repartition(num_partitions) # Adjust Spark configurations if record_count < LARGE_DATA_THRESHOLD: spark.conf.set("spark.sql.shuffle.partitions", str(max(1, num_partitions))) else: spark.conf.set("spark.sql.shuffle.partitions", "200") spark.conf.set("spark.sql.files.maxRecordsPerFile", "10000000") spark.conf.set("spark.sql.parquet.mergeSchema", "false") spark.conf.set("spark.sql.parquet.filterPushdown", "true") # Write data to output path df.write.format("parquet") \ .option("batchsize", "10000") \ .option("socketTimeout", "600000") \ .option("connectTimeout", "60000") \ .mode("overwrite").save(output_blob_path) print(f"{datetime.now()} Table '{table_name}' has been cloned in Azure Blob storage successfully:{output_blob_path}") except Exception as e: error_message = str(e) raise
Masking Process
Azure Databricks Job for Database constraints
def collect_database_script_single(script_query, script_file_name, operations_timestamp): # connection details user = dbutils.secrets.get(scope="dm", key="mss-oracle-user") password = dbutils.secrets.get(scope="dm", key="mss-oracle-password") jdbcHostname = dbutils.secrets.get(scope="dm", key="mss-oracle-hostname") jdbcDatabase = dbutils.secrets.get(scope="dm", key="mss-oracle-database") jdbcPort = 1521 # Step 1: Define Oracle JDBC connection properties jdbc_url = f"jdbc:oracle:thin:@{jdbcHostname}:{jdbcPort}/{jdbcDatabase}" connection_properties = { "user": user, "password": password, "driver": "oracle.jdbc.OracleDriver" } # Step 2: Execute the query using JDBC and collect the result df = spark.read.jdbc(url=jdbc_url, table=f"({script_query})", properties=connection_properties) # Step 3: Collect the result into a list of strings sql_statements = df.collect() # Step 4: Prepare the final SQL script as a string sql_script = "\n".join([row[0] for row in sql_statements]) # Step 5: Define the file path in mounted storage output_file_path = f"{MOUNTPOINT}/mss/operations/{operations_timestamp}/scripts/{script_file_name}" # Step 6: Write the SQL script to the file in Azure Blob Storage dbutils.fs.put(output_file_path, sql_script, overwrite=True) print(f"SQL script saved to {output_file_path}")
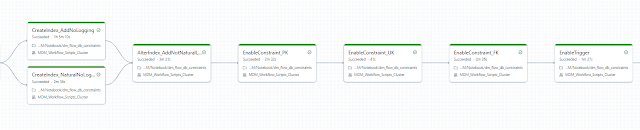
- DisableTrigger: This step is responsible for disabling all triggers in the database.
- DisableConstraint_FK: This job focuses on disabling all foreign key constraints.
- DisableConstraint_UK: This job handles the disabling of unique constraints. However, not all unique constraints were disabled—only those used in primary key constraints.
- DisableConstraint_PK: This job disables primary key constraints.
- DropIndex: The final job in the sequence drops or disables indexes. Different database platforms offer varying levels of optimization for handling triggers. For example, disabling triggers in Oracle databases can be challenging due to the platform's archaic structure. Adding new SQL DDL capabilities in Oracle often faces resistance from its foundational design and the philosophy of its original creators.
def execute_oracle_sql_script_jaydebeapi(operations_timestamp, script_file_name, jdbc_url, username, password): # Step 1: Define file paths sql_file_path = f"{MOUNTPOINT}/mss/operations/{operations_timestamp}/scripts/{script_file_name}" log_file_name_part = os.path.splitext(script_file_name)[0] log_file_path = f"{MOUNTPOINT}/mss/operations/{operations_timestamp}/logs/sql_execution_log_{log_file_name_part}_{datetime.now().strftime('%Y%m%d_%H%M%S')}.log" # Step 2: Read the SQL script from file df = spark.read.text(sql_file_path) lines = df.collect() # Collect the DataFrame rows (lines of the SQL script) # Step 3: Parse SQL statements by splitting them by semicolons sql_statements = [] current_statement = [] for row in lines: line = row['value'].strip() current_statement.append(line) if line.endswith(";"): # End of an SQL statement full_statement = " ".join(current_statement) # Combine parts into a full statement sql_statements.append(full_statement) # Add it to the list current_statement = [] # Reset for next statement # Step 4: Split the SQL statements into chunks using numpy (for parallel execution) num_chunks = 8 # Number of parallel chunks sql_chunks = np.array_split(sql_statements, num_chunks) # Step 5: Prepare (chunk, chunk_id) tuples for parallel execution chunk_with_ids = [(chunk, idx) for idx, chunk in enumerate(sql_chunks)] # Define the function to execute a list of SQL statements and return logs from each worker def execute_sql_list_and_collect_logs(sql_list_with_id): sql_list, chunk_id = sql_list_with_id # Unpack the tuple (list of SQLs, chunk ID) worker_logs = [] # Collect logs in a local list for each worker # Add an iteration ID to track the execution of each statement for idx, sql in enumerate(sql_list, start=1): # enumerate adds the iteration ID starting from 1 try: # Remove any semicolons from the SQL line sql_statement = sql.replace(';', '') # Execute the SQL statement using the provided function sql_log_message = execute_oracle_ddl_statement_jaydebeapi(sql_statement, jdbc_url, username, password) # Add iteration ID and log message log_message = f"{datetime.now()} - Chunk {chunk_id}, Iteration {idx} - Executed: {sql_log_message}" worker_logs.append(log_message) # Collect the log message except Exception as e: # Collect error message, including iteration ID log_message = f"{datetime.now()} - Chunk {chunk_id}, # {idx} - Error executing: {sql_statement[:50]} - {str(e)}" worker_logs.append(log_message) # Collect the log message return worker_logs # Return the collected logs from each worker # Step 6: Run the chunks in parallel using sc.parallelize() rdd = sc.parallelize(chunk_with_ids, numSlices=num_chunks) # Collect logs from all workers collected_logs = rdd.flatMap(execute_sql_list_and_collect_logs).collect() # Step 7: Write all collected logs to the log file if collected_logs: log_content = "\n".join(collected_logs) dbutils.fs.put(log_file_path, log_content, overwrite=True) print(f"Logs for {log_file_name_part} are written to {log_file_path}") else: print("No logs were collected.") # Example call to the function (replace with actual values) # execute_oracle_sql_script_jaydebeapi("202410062107", "DisableIndex_TEST.sql", jdbc_url, username, password)
def execute_oracle_ddl_statement_jaydebeapi(sql_statement, jdbc_url, username, password): try: # Connect without JPype using pure JDBC mode connection = jaydebeapi.connect( "oracle.jdbc.OracleDriver", # JDBC driver class jdbc_url, # JDBC URL for Oracle [username, password], # Credentials "/Workspace/Shared/MDM/Library/ojdbc8.jar" # Path to the JDBC JAR ) connection.jconn.setAutoCommit(False) # Disable auto-commit cursor = connection.cursor() # Create a cursor object cursor.execute(sql_statement) # Execute the DDL statement connection.commit() # Commit the transaction return f"Success: DDL statement: {sql_statement[:100]}" except Exception as e: if 'connection' in locals(): connection.rollback() return f"Failed: DDL statement: {sql_statement[:100]}, Error: {str(e)}" finally: if 'cursor' in locals(): cursor.close() if 'connection' in locals(): connection.close()
- CreateIndex: The first step in the sequence recreates or enables all indexes. Indexes are reintroduced to optimize query performance and ensure efficient database operations.
- EnableConstraint_PK: This job restores primary key constraints, re-establishing the unique identification of rows within each table.
- EnableConstraint_UK: This step enables unique constraints that were previously disabled, ensuring the integrity of columns requiring unique values. Only those associated with primary key constraints are re-enabled.
- EnableConstraint_FK: This job restores all foreign key constraints, re-establishing the relationships between tables and ensuring referential integrity.
- EnableTrigger: The final step in the sequence re-enables all triggers, restoring any automated database behaviours that respond to specific events.
Azure Databricks Job for Masking
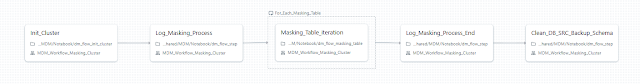
The job responsible for running the masking process on all required databases is called by its parent job but can also be executed independently. While including all the necessary initialization and logging steps, the purpose of this job is straightforward: it takes a table name as an input parameter and passes it further to execute all the required steps for the table (data copy, pre-processing validation, masking, and post-processing validation). These steps are performed in parallel.
Job parameters
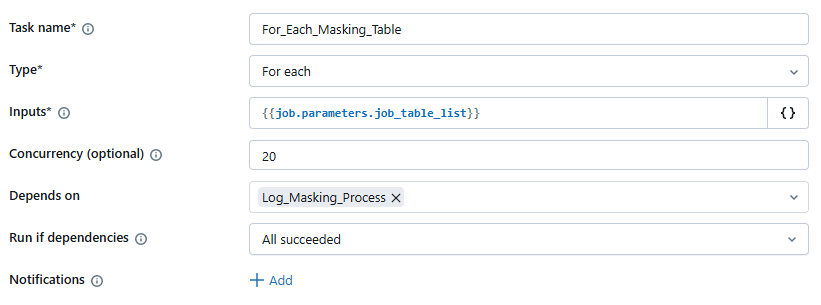
For Each Masking Table iteration
Then, each individual element of the {{input}} array is passed to the dm_flow_masking_table Python notebook to sequentially execute the following steps: (1) data copy, (2) pre-processing validation, (3) masking, and (4) post-processing.
Python dm_flow_masking_table Notebook
# Split `table_name` into `schema_name` and `table_name_only` schema_name, table_name_only = table_name.split(".") dm_databricks_workspacename = dbutils.secrets.get(scope="dm", key="dm-databricks-workspacename") df = reading_dm_config_table(dm_databricks_workspacename) # Filter the DataFrame and select the required columns, keeping it as a DataFrame df_target_table_metadata = df.filter( (df["schema_name"] == schema_name) & (df["table_name"] == table_name_only) ) # Extract the values for `database_name` and `host_port` row = df_target_table_metadata.first() database_name = row["database_name"] if row is not None else None host_port = str(row["port"]) table_name_source = f"{schema_name}_src.{table_name_only}" # Uninitialized DataFrames df_target_table_data = None df_sourcing_table_data = None if 'backup' in operations_list: print(f"{datetime.now()} Table '{table_name}' backup started") # Read target table into DataFrame df_target_table_data = dm_read_table_into_dataframe(database_name, table_name, host_port) dm_flow_table_backup( database_name, table_name, operations_timestamp, "['file_storage', 'database']", df_target_table_data, df_target_table_metadata ) if 'validation_begin' in operations_list: print(f"{datetime.now()} Table '{table_name}' validation started") update_masking_table_status(table_name, str(operations_timestamp), "PreValidation", "Started") # Read target table into DataFrame if df_target_table_data is None or not df_target_table_data.schema.fields: df_target_table_data = dm_read_table_into_dataframe(database_name, table_name, host_port) # Read sourcing table into DataFrame df_sourcing_table_data = dm_read_table_into_dataframe(database_name, table_name_source, host_port) dm_flow_table_validation( database_name, table_name, 'begin', df_target_table_data, df_target_table_metadata, df_sourcing_table_data ) update_masking_table_status(table_name, str(operations_timestamp), "PreValidation", "Completed") if 'masking' in operations_list: print(f"{datetime.now()} Table '{table_name}' masking started") update_masking_table_status(table_name, str(operations_timestamp), "MaskingStatus", "Started") # Read sourcing table into DataFrame if df_sourcing_table_data is None or not df_sourcing_table_data.schema.fields: df_sourcing_table_data = dm_read_table_into_dataframe(database_name, table_name_source, host_port) df_masked_table_data = dm_flow_table_masking( database_name, table_name, df_sourcing_table_data, df_target_table_metadata ) print(f"{datetime.now()} df_masked_table_data '{df_masked_table_data.count()}' records.") update_masking_table_status(table_name, str(operations_timestamp), "MaskingStatus", "Completed") if 'validation_end' in operations_list: print(f"{datetime.now()} Table '{table_name}' validation started") update_masking_table_status(table_name, str(operations_timestamp), "PostValidation", "Started") # Read target table into DataFrame if df_masked_table_data is None or not df_masked_table_data.schema.fields: df_masked_table_data = dm_read_table_into_dataframe(database_name, table_name, host_port) # Read sourcing table into DataFrame if df_sourcing_table_data is None or not df_sourcing_table_data.schema.fields: df_sourcing_table_data = dm_read_table_into_dataframe(database_name, table_name_source, host_port) print(f"{datetime.now()} df_masked_table_data '{df_masked_table_data.count()}' records.") print(f"{datetime.now()} df_sourcing_table_data '{df_sourcing_table_data.count()}' records.") dm_flow_table_validation( database_name, table_name, 'end', df_masked_table_data, df_target_table_metadata, df_sourcing_table_data ) update_masking_table_status(table_name, str(operations_timestamp), "PostValidation", "Completed")
Python dm_flow_table_masking Function
def dm_flow_table_masking(database_name, target_table_name, df_target_table_data, df_target_table_metadata): try: run_logging_to_blob_history('Info', f'dm_flow_table_masking({database_name}:{target_table_name})', None, None, None, None, None, 'Started') source_table_name = f"{target_table_name.split('.')[0]}_src.{target_table_name.split('.')[1]}" print(f"Tables to mask: {target_table_name} from {source_table_name}.") mss_user, mss_password, jdbcUrl, jdbc_driver_class = dm_oracle_connection() current_table_data_df = df_target_table_data print(f"current_table_data_df Table: {current_table_data_df.count()}...") current_table_metadata_df = df_target_table_metadata.select("column_name", "masking_rule", "masking_type") source_schema, source_table = source_table_name.split('.') for row in current_table_metadata_df.collect(): column_name, masking_rule, masking_type = row["column_name"], row["masking_rule"], row["masking_type"] width = get_column_width(jdbcUrl, mss_user, mss_password, source_table_name, column_name) if masking_rule == "data_redaction_simple": current_table_data_df = current_table_data_df.withColumn("masking_type_temp", lit(masking_type)) current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_redaction_simple_udf(col(column_name), col("masking_type_temp"), lit(width)) ).drop("masking_type_temp") current_table_data_df = current_table_data_df.drop("masking_type_temp") elif masking_rule == "data_randomization": current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_randomization_udf(col(column_name).cast("string"), lit(masking_type)) ) elif masking_rule == "data_nullificaion": current_table_data_df = current_table_data_df.withColumn( column_name, lit(None).cast(current_table_data_df.schema[column_name].dataType) ) elif masking_rule == "data_lookup_substitution_name_simple": current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_substitution_name_simple_udf(col(column_name), lit(width), lit(masking_type)) ) elif masking_rule == "data_lookup_substitution_address_street": current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_substitution_address_street_udf(col(column_name), lit(width)) ) elif masking_rule == "data_lookup_substitution_address_other": current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_substitution_address_other_udf(col(column_name)) ) elif masking_rule == "data_lookup_substitution_employer": current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_substitution_employer_udf(col(column_name), lit(width)) ) elif masking_rule == "data_lookup_substitution_fullname": current_table_data_df = current_table_data_df.withColumn( column_name, dm_masking_rule_data_substitution_fullname_udf(col(column_name), lit(width)) ) elif masking_rule == "data_lookup_substitution_city_postal_code": column_name = [item.strip() for item in column_name[1:-1].split(',')] if masking_type == 'postalcode': columnname = column_name[1] else: #CITY name case columnname = column_name[0] if len(column_name) ==3: #case when we have a complete set of attributes (City, Postal Code, Province/State) current_table_data_df = current_table_data_df.withColumn( columnname, dm_masking_rule_data_substitution_city_postalcode_province_udf(col(column_name[0]), col(column_name[1]), lit(masking_type), col(column_name[2])) ) else: # otherwise City, Masking Type, Province/State current_table_data_df = current_table_data_df.withColumn( columnname, dm_masking_rule_data_substitution_city_postalcode_province_udf(col(column_name[0]), lit(masking_type),col(column_name[1])) ) run_logging_to_blob_history( 'Info', f'DataMasking Process Started on {source_table_name}', 'Data Masking Operation', database_name, source_schema, source_table, None, 'Started' ) # Writing masked data to the original target table backup_table_name = f"{target_table_name.split('.')[0]}_src.{target_table_name.split('.')[1]}" dm_append_dataframe_to_table(current_table_data_df, target_table_name, backup_table_name, jdbcUrl, mss_user, mss_password) run_logging_to_blob_history( 'Info', f'DataMasking Process Completed on {source_table_name} from {target_table_name}', 'Data Masking Operation', database_name, source_schema, source_table, None, 'Completed' ) return current_table_data_df except Exception as e: error_message = str(e) run_logging_to_blob_history( 'Error', f"dm_flow_table_masking({database_name}) : {error_message}", 'Data Masking Operation', database_name, None, source_table_name, None, 'Failed' ) run_logging_to_sql_history() raise
Masking logic explained
The simplicity, beauty, and elegance of this meta-driven solution for table data masking, data transformation, and data obfuscation lie in its design. The entire set of tables for masking is processed dynamically. Each table has a unique set of columns requiring masking, and each column is associated with a specific masking rule.
Pre- and post-validation were not that complicated, primarily checking table schema consistency and data changes at the end. These steps were less important since the most critical part was ensuring that the masked data changes were successfully written to persistent storage. The complete Databricks solution included 13 Python notebooks with over a hundred Python functions, ranging from simple to complex. At the end of the day, it proved that Azure Databricks can be used for a metadata-driven masking process, and it can do this job really well!








Comments
Post a Comment